COVID-19 - Ireland County Data
Introduction
The Irish government has made available several datasets relating to the COVID-19 pandemic. These are available at the Ireland’s COVID-19 Data Hub.
This example looks at how you can download and produce visualisations of the County Statistics datasets (Covid19CountyStatisticsHPSCIreland) using a Python script.
Libraries
The following libraries are used in the script:
- argparse - makes it easy to write user-friendly command-line interfaces buy handling command line parameters.
- requests - allows sending HTTP requests through Python, which will be used to obtain the dataset from the internet.
- pandas - open-source Python library that provides powerful data structures and data analysis tools to deal with datasets.
- matplotlib.pyplot - used for data visualisation.
- matplotlib.dates - used for date formatting on data visualisations.
import argparse
import requests
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.dates as mdatesCommand Line Arguments
The scripts takes two parameters:
- county - the name of the county you want to plot data for
- days - the number of days data you want to plot
# Get command line arguments
parser = argparse.ArgumentParser()
parser.add_argument("--county", help="county name - default: Cork", default="Cork")
parser.add_argument("--days", help="number of days to display data for - default: 30", type=int, default="30")
args = parser.parse_args()Get Dataset
The CSV version of the dataset is downloaded to the local file system.
# Get latest county date in csv format
filename = 'covid-ie-counties.csv'
data_url = 'https://opendata-geohive.hub.arcgis.com/datasets/d9be85b30d7748b5b7c09450b8aede63_0.csv?outSR=%7B%22latestWkid%22%3A3857%2C%22wkid%22%3A102100%7D'
data_content = requests.get(data_url).content
csv_file = open(filename, 'wb')
csv_file.write(data_content)
csv_file.close()Filter Dataset
The CSV file is loaded into a pandas DataFrame and filtered by county name and the number of days to plot.
# Filter data by county and number of days
df = pd.read_csv(filename)
df = df.loc[df['CountyName'] == args.county].tail(args.days)Format Date
When loaded into the dataframe the TimeStamp is a string, convert it into a datetime object for charting and formatting.
# Converting time stamp to a to datetime e.g. 2020/03/22 00:00:0
df['TimeStamp'] = pd.to_datetime(df['TimeStamp'], format='%Y/%m/%d %H:%M:%S')Bar Chart - Cumulative Confirmed Cases By County
The cumulative confirmed cases are plotted using the TimeStamp and ConfirmedCovidCases fields in the DataFrame.
# Bar Chart - Cumulative Confirmed Cases By County
plt.bar(df["TimeStamp"], df["ConfirmedCovidCases"])
plt.xlabel('Date')
plt.ylabel('Total Cases')
plt.title('Cumulative Confirmed Cases for ' + args.county + ' for last ' + str(args.days) + ' days')
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%d-%b'))
plt.gcf().autofmt_xdate()
plt.show()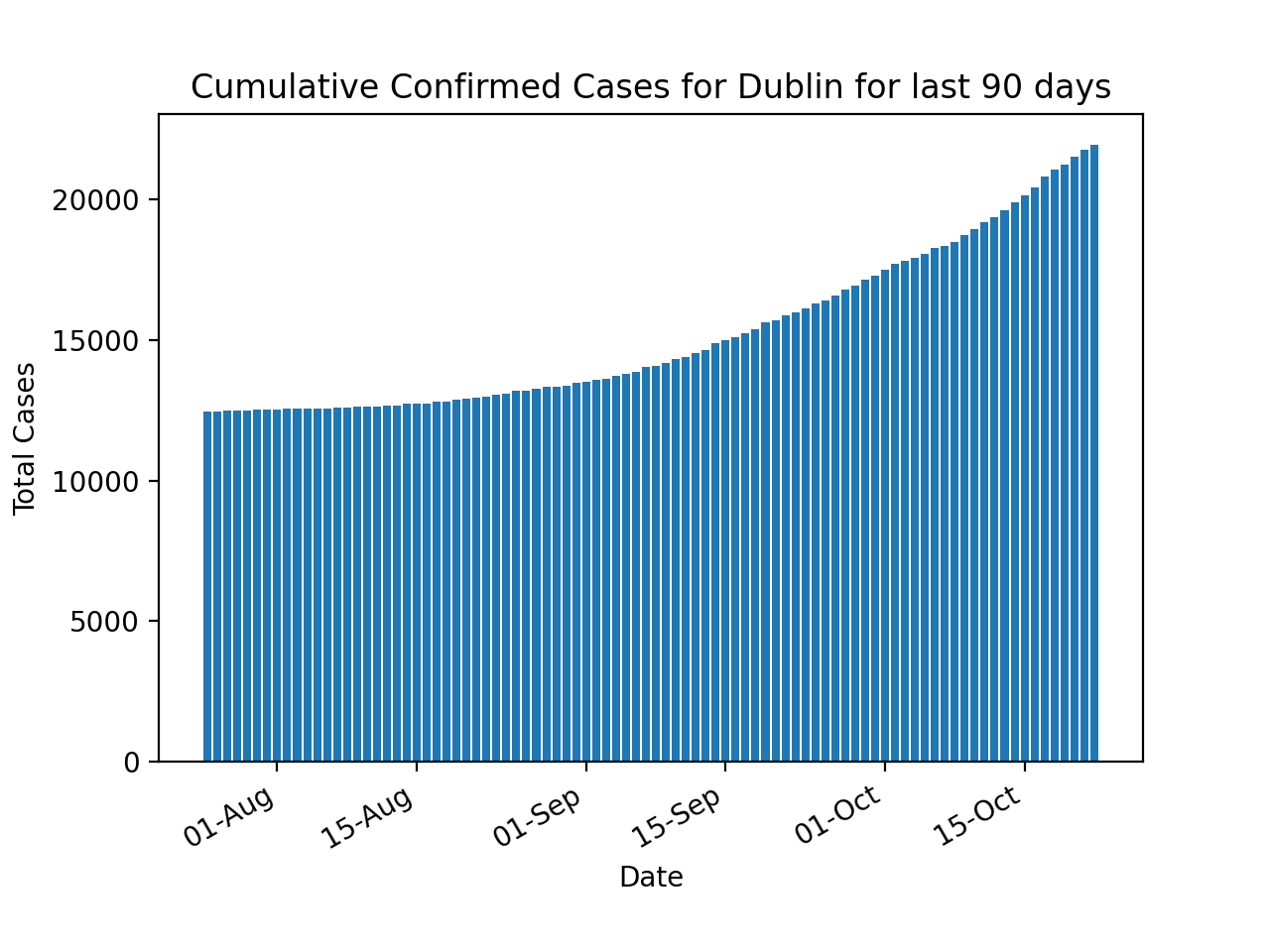
Bar Chart - New Confirmed Cases By County
The new daily confirmed cases are plotted using the TimeStamp and the diff method on ConfirmedCovidCases. The diff method calculates the difference of a DataFrame element compared with another element in the DataFrame (default is element in previous row).
# Bar Chart - New Confirmed Cases By County
plt.bar(df["TimeStamp"], df["ConfirmedCovidCases"].diff())
plt.xlabel('Date')
plt.ylabel('Confirmed Cases')
plt.title('New Confirmed Cases for ' + args.county + ' for last ' + str(args.days) + ' days')
plt.gca().xaxis.set_major_formatter(mdates.DateFormatter('%d-%b'))
plt.gcf().autofmt_xdate()
plt.show()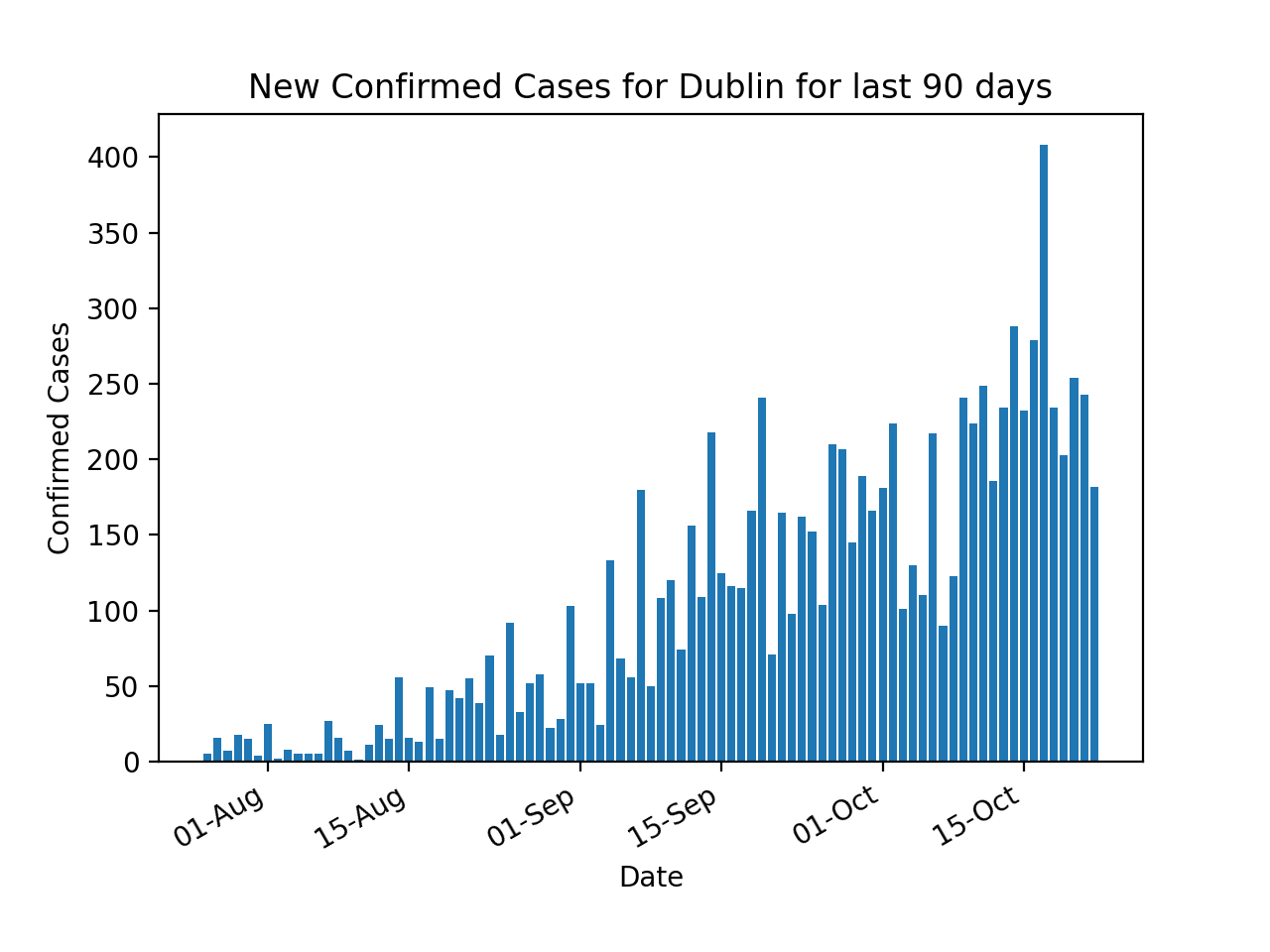
Usage
Help for parameters can be viewed by:
$ python3 covid-ie-counties.py --help
usage: covid-ie-counties.py [-h] [--county COUNTY] [--days DAYS]
optional arguments:
-h, --help show this help message and exit
--county COUNTY county name - default: Cork
--days DAYS number of days to display data for - default: 30This is an example of the command to plot the previous 30 days of data for county Dublin:
$ python3 covid-ie-counties.py --county Dublin --days 30Conclusion
This is just one example of what can be done with the datasets available from the Ireland’s COVID-19 Data Hub.
Full code for this example available on GitHub here.
